A few months ago, I wrote a post about junk DNA and ENCODE. Since then, more evidence has surfaced so I’ve decided to make this into a blog post. I’ve slightly modified the original post as well as added the new information plus all the relevant links. This is a long and sometimes technical post. Note: “Creationist” is interchangeable with ID-proponent. They’re the same.
The main problem with this story is not what scientists have claimed and then found, but rather what the popular press has (mis)understood. This is also a story about scientists failing to communicate science properly. AronRa said on the 31st of May, 2009, in his video Ida Know (the first of a five-part summary about the 47-million year old primate fossil Ida) the following, which also holds true about this story:
But sadly, the media isn’t entirely to blame, some of this has been done by scientists.
…
It is highly inappropriate sensationalism and the way it’s described is very misleading to anybody who doesn’t understand taxonomy very well and almost nobody does.
The same is true in this case, only substitute “taxonomy” with “evolution”, “genetics” and “biochemistry”.
A pop-science journal Arstechnica (Author John Timmer) has also commented on this phenomenon:
ArsTechnica: Most of what you read was wrong: how press releases rewrote scientific history
Many press reports that resulted [from the ENCODE release] painted an entirely fictitious history of biology’s past, along with a misleading picture of its present. As a result, the public that relied on those press reports now has a completely mistaken view of our current state of knowledge (this happens to be the exact opposite of what journalism is intended to accomplish). But you can’t entirely blame the press in this case. They were egged on by the journals and university press offices that promoted the work,and, in some cases, the scientists themselves.
…
Unfortunately, things like well-established facts make for a lousy story. So instead, the press has often turned to myths, aided and abetted by the university press offices and scientists that should have been helping to make sure they produced an accurate story.
I’ll go into the details of the ENCODE story near the end, but first we need a short history of “Junk DNA”.
A history of “junk DNA”
The story begins with Susumu Ohno. In 1970, he wrote a book (Ohno, 1970a ) in which he laid out the argument for the role of gene duplication in evolution. We now know that it does indeed play its part. One thought experiment he had regarded genes that were duplicated (in his example, three sequences sharing the same sequence) and not under pressure by selection any more. If there is no selective pressure, they would mutate and two out of three would likely serve no function, due to high mutation rates.
Ohno 1970a, p.62
[It is likely that] in a relatively short time, two of the three duplicates would join the ranks of ‘garbage DNA’.
This was the first time anything like this was proposed. Only two years later, in another paper (Ohno, 1972) would he coin the phrase “junk DNA”. So what was “junk DNA” or “garbage DNA” to him? Well it’s based on a very well-known observation:
Ohno 1972
If we take the simplistic assumption that the number of genes contained is proportional to the genome size, we would have to conclude that 3 million or so genes are contained in our genome. The falseness of such an assumption becomes clear when we realize that the genome of the lowly lungfish and salamanders can be 36 times greater than our own.
As we now know, we have roughly 20,000 genes, which fit well with Ohno’s prediction of no more than 30,000 genes. It was also observed that there can be a lot of duplications and insertion of retroposons without affecting the body in any way. I talk about this later on, under the heading “pseudogenes”.
At the time of Ohno’s writing, “junk DNA” was “meant to describe the loss of protein-coding function by deactivated gene duplicates, which in turn were believed to constitute the bulk of eukaryotic genomes”. (Genomicron, 2007)
A very important part follows:
As different types of non-coding DNA were identified, the concept of gene duplication as their source, and therefore “junk DNA” as their descriptor, found new and broader application. However, it is now clear that most non-coding DNA is not produced by this mechanism, and is therefore not accurately described as “junk” in the original sense.
So in the original sense, we don’t have a lot of “junk DNA” after all. The important thing to know here is that the term has been butchered by the media to mean all non-coding DNA, which strictly shouldn’t be called “junk DNA” but rather “pseudogene”, coined in 1977 (Jacq et al. 1977) to describe a functionless gene. Now note the miscommunication: There is a difference between “junk” (stuff one keeps) and “trash” (stuff one throws out). This was noted in 1988:
Brenner 1998
There is the rubbish we keep, which is junk, and the rubbish we throw away, which is garbage.
And in 1990, Brenner said the following:

(S. Brenner, The human genome: the nature of the enterprise (in: Human Genetic Information: Science, Law and Ethics, No. 149: Science, Law and Ethics, Symposium Proceedings (CIBA Foundation Symposia) John Wiley and Sons Ltd 1990, Source) <– One problem with that blog post is that much of it is wrong. I merely provide the source to show where I got the picture from.
And even in 1973, Ohno suggested a potential function for “junk DNA”:
Ohno, 1973
The bulk of functionless DNA in the mammalian genome may serve as a damper to give a reasonably long cell generation time (12 hours or so instead of several minutes)
Genomicron, 2007
From the very beginning, the concept of “junk DNA” has implied non-functionality with regards to protein-coding, but left open the question of sequence-independent impacts (perhaps even functions) at the cellular level. “Junk DNA” may now be taken to imply total non-function and is rightly considered problematic for that reason, but no such tacit assumption was present in the term when it was coined.
Gregory goes on to make a very astute observation: If there is no function for all genes, creationists are in serious trouble. (Note: Recent reading of a creationist blog post suggests that there is at least one creationist who does not adhere to this and thinks it wouldn’t matter much if their prediction weren’t true. I’ve yet to find the original source [a guy called Axe?] so I’m left to wonder how that should work… This doesn’t detract from the point that most creationists do hold the view presented both above and below.)
Genomicron, 2007
[This is why] all non-coding DNA must, a priori, be functional.
To satisfy this expectation, creationist authors (borrowing, of course, from the work of molecular biologists, as they do no such research themselves) simply equivocate the various types of non-coding DNA, and mistakenly suggest that functions discovered for a few examples of some types of non-coding sequences indicate functions for all (see Max 2002 for a cogent rebuttal to these creationist confusions). Case in point: a few years ago, much ado was made of Beaton and Cavalier-Smith’s (1999) titular proclamation, based on a survey of cryptomonad nuclear and nucleomorphic genomes, that “eukaryotic non-coding DNA is functional”. The point was evidently lost that the function proposed by Beaton and Cavalier-Smith (1999) was based entirely on coevolutionary interactions between nucleus size and cell size.
Apart from the above mentioned potential function for “junk DNA”, many more have been identified since:
Genomicron, 2007
Examples include buffering against mutations (e.g., Comings 1972; Patrushev and Minkevich 2006) or retroviruses (e.g., Bremmerman 1987) or fluctuations in intracellular solute concentrations (Vinogradov 1998), serving as binding sites for regulatory molecules (Zuckerkandl 1981), facilitating recombination (e.g., Comings 1972; Gall 1981; Comeron 2001), inhibiting recombination (Zuckerkandl and Hennig 1995), influencing gene expression (Britten and Davidson 1969; Georgiev 1969; Nowak 1994; Zuckerkandl and Hennig 1995; Zuckerkandl 1997), increasing evolutionary flexibility (e.g., Britten and Davidson 1969, 1971; Jain 1980; reviewed critically in Doolittle 1982), maintaining chromosome structure and behaviour (e.g., Walker et al. 1969; Yunis and Yasmineh 1971; Bennett 1982; Zuckerkandl and Hennig 1995), coordingating genome function (Shapiro and von Sternberg 2005), and providing multiple copies of genes to be recruited when needed (Roels 1966).
In addition, I believe one can add both Epigenetics and Evo-Devo to that list.
Finally, Genomicron notes the following:
Genomicron, 2007
More broadly, those who would attribute a universal function for non-coding DNA must bear the following in mind: any proposed function for all non-coding DNA must explain why an onion or a grasshopper needs five times more of it than anyone reading this sentence.
Pseudogenes
Now I need to explain pseudogenes. I think the easiest way is to use this picture from the wikipedia article, which I modified for the purpose of illustration:
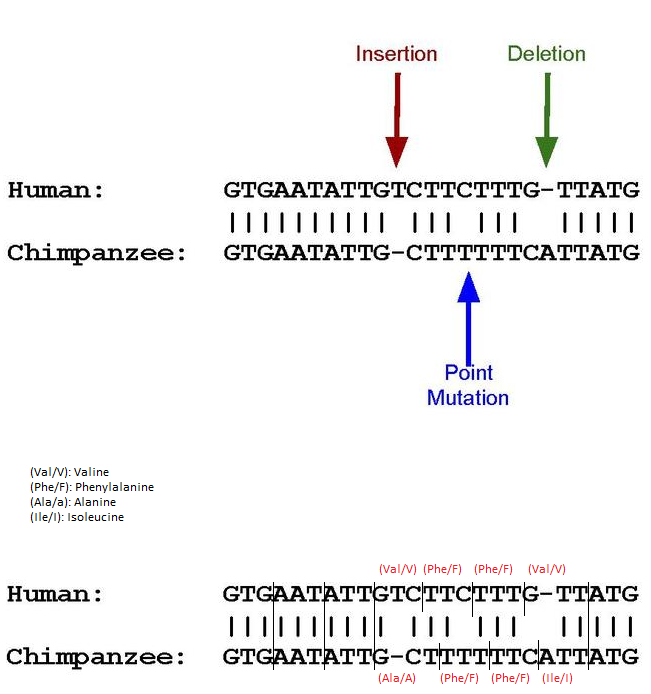
As you may know, amino acids are encoded by reading DNA sequences in triplets. If, as in the above sequence, an insertion, deletion and point mutation occurs, the triplets are read differently. The results in amino acid encoding are shown above. If the new sequences produce premature stop-codons or, as in this case, simply different amino acids, genes may not be activated and proteins may not be produced. Sometimes they are encoded but do not actively help the organism.
These are then called pseudogenes.
The ENCODE delusion
A few months ago, the ENCODE staff published some research, which prompted the following statement by the EFF:
On September 19, the Ninth Circuit is set to hear new arguments in Haskell v. Harris, a case challenging California’s warrantless DNA collection program. Today EFF asked the court to consider ground-breaking new research that confirms for the first time that over 80% of our DNA that was once thought to have no function, actually plays a critical role in controlling how our cells, tissue and organs behave.
But as I showed above, functions for “junk DNA” have been known since before the term was even coined! This is what I’m talking about when criticizing the way scientists convey science and the way newspapers bring it to the public. (Luckily, some scientists have spoken out against the ENCODE fiasco. And hey, even some folk from the ID-crowd.)
But let’s back up a little. ENCODE has been working for quite some time now and, as you would expect, it has been talked about since at least 2007. (Arthur Hunt on Panda’s Thumb, 2007) Even then, Creationists wanted to claim what they claim now, namely that all DNA has a definite function. There was (at least) one problem for them: A 2005 paper (Wyers et al. 2005) showed that “much of the RNA made by a cell is thrown away. This includes RNA encoded by intergenic regions.” (Quote from Arthur Hunt, 2007)
PZ Myers documents a second story, with a 2010 paper (van Bakel et al. 2010) explaining once again that genes only make up about 2% of the genome, while the rest is non-coding.
That takes us back to the 2012 report from ENCODE. They claimed that 80% of the genome serves some biochemical function, with “function” being defined as participating “in at least one biochemical RNA- and/or chromatin-associated event in at least one cell type”. That’s where the real problem in communication lies: The definition of the word “function”.
That isn’t function. That isn’t even close. And it’s a million light years away from “a critical role in controlling how our cells, tissue and organs behave”. All that says is that any one bit of DNA is going to have something bound to it at some point in some cell in the human body, or may even be transcribed. This isn’t just a loose and liberal definition of “function”, it’s an utterly useless one.
I’ll try to make this as clear as possible, so I’ll draw on an analogy. Imagine you found two ball-point pens. One of them is a normal pen, the other lacks the ball-point. The first pen has a definite function: To write. The second one may have other functions (like removing ear-wax from your ear), but that’s not a definition of function any sane person would recognize.
The same happened to ENCODE: The way they define “function” is so broad that it’s absolutely useless. Indeed, much randomly generated DNA can be said to have “function” under this definition.
Also note that in his blog, Ewan Birney (kind of) explains what “function” means in ENCODE terms. What’s noticeable is that it could just as well have been swapped for “specific biochemical activity”, which takes the wind out of the sails completely.
In another Q&A point, he said that using the 80% number instead of the 20% functional bases (notice a difference?) was used to… hype up the story. That’s basically it.
Ewan Birney, Q&A
Q. Ok, fair enough. But are you most comfortable with the 10% to 20% figure for the hard-core functional bases? Why emphasize the 80% figure in the abstract and press release?
A. (Sigh.) Indeed. Originally I pushed for using an “80% overall” figure and a “20% conservative floor” figure, since the 20% was extrapolated from the sampling. But putting two percentage-based numbers in the same breath/paragraph is asking a lot of your listener/reader, they need to understand why there is such a big difference between the two numbers, and that takes perhaps more explaining than most people have the patience for. We had to decide on a percentage, because that is easier to visualize, and we choose 80% because (a) it is inclusive of all the ENCODE experiments (and we did not want to leave any of the sub-projects out) and (b) 80% best coveys the difference between a genome made mostly of dead wood and one that is alive with activity. We refer also to “4 million switches”, and that represents the bound motifs and footprints.We use the bigger number because it brings home the impact of this work to a much wider audience. But we are in fact using an accurate, well-defined figure when we say that 80% of the genome has specific biological activity.
And again from Arstechnica:
So even as the [2007] paper was released, we already knew the ENCODE definition of “functional impact” was, at best, broad to the point of being meaningless. At worst, it was actively misleading.
…
In the lead paper of a series of 30 released this week, the ENCODE team decided to redefine “functional.” Instead of RNA, its new definition was more DNA focused, and included sequences that display “a reproducible biochemical signature (for example, protein binding, or a specific chromatin structure).” In other words, if a protein sticks there or the DNA isn’t packaged too tightly to be used, then it was functional.
That definition nicely encompasses the valuable regulatory DNA, which controls nearby genes through the proteins that stick to it. But,and this is critical,it also encompasses junk DNA. Viruses and transposons have regulatory DNA to ensure they’re active; genes can pick up mutations in their coding sequence that leave their regulatory DNA intact. In short, junk DNA would be expected to include some regulatory DNA, and thus appear functional by ENCODE’s definition.
…
The ENCODE team itself bears a particular responsibility here. The scientists themselves should have known what the most critical part of the story was,the definition of “functional” and all the nuance and caveats involved in that,and made sure the press officers understood it. Those press officers knew they would play a key role in shaping the resulting coverage, and should have made sure they got this right. The team has now failed to do this twice.
All that being said, Sandwalk notes two things:
1) The word the ENCODE-people are looking for is not “function”, it’s “noise“.
2) The debate isn’t only about the definition of “function”, but something deeper. An additional problem may be that some scientists don’t understand evolution. That’s pretty sad in this day and age.
A further update comes from a 2013 paper in “Genome Biology and Evolution”. The paper is discussed over at Pharyngula and it basically rips into ENCODE’s papers. There’s a lot of technical stuff I needn’t cover, so I’ll limit myself to mentioning one thing: Other researchers have found only 10% true functionality, that’s 70% less than the folk over at ENCODE.
I’ll conclude:
1) “Junk” DNA was, from the moment of its conception, a misnomer. Just like “Big Bang” falsely conjures the image of an explosion, “junk DNA” falsely conjures the image of complete non-function or garbage. However, in both cases that’s not consistent with what scientists have been saying even before the term was coined. “Junk DNA” should not be in common usage. A more precise term would be “pseudogene”.
2) Conveying what “junk DNA”, non-coding DNA and pseudogenes are, as well as the nuanced differences between the terms, is a difficult job. Sadly, neither scientists nor journalists have done a good job of explaining the terms. It would be interesting to make an extremely careful and detailed YT series on this subject.
3) Much of the ENCODE hype rests on the definition of the term “function”. If the general public and creationists were aware of what “function” means in ENCODE terms, the hype would almost completely fade away. Note that I’m not saying that the results were wrong, inconclusive or not worthy of recognition, I’m simply saying that they were over-hyped. (Which would put them into the second circle of scientific hell!)
4) This problem also creates an opportunity. We now understand what we did wrong and this may encourage scientists to be more careful in the future when explaining things. I hope to make a blog series on the public understanding of science soon and this will be one of my focuses.
Below are all the references used in the creation of this post. The first one is only scientific papers, the second one is blog posts and opinion pieces.
All references with links attached are the resources I used myself, other resources in plain black are additional resources.
References
Andolfatto, P. 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149-1152.
Batten, D. 1998. ‘Junk’ DNA (again). Creation Ex Nihilo Technical Journal 12: 5.
Beaton, M.J. and T. Cavalier-Smith. 1999. Eukaryotic non-coding DNA is functional: evidence from the differential scaling of cryptomonad genomes. Proceedings of the Royal Society of London, Series B 266: 2053-2059.
Bejerano, G., M. Pheasant, I. Makunin, S. Stephen, W.J. Kent, J.S. Mattick, and D. Haussler. 2004. Ultraconserved elements in the human genome. Science 304: 1321-1325.
Bennett, M.D. 1982. Nucleotypic basis of the spatial ordering of chromosomes in eukaryotes and the implications of the order for genome evolution and phenotypic variation. In Genome Evolution (eds. G.A. Dover and R.B. Flavell), pp. 239-261. Academic Press, New York.
Bergman, J. 2001. The functions of introns: from junk DNA to designed DNA. Perspectives on Science and Christian Faith 53: 170-178.
Biémont, C. and C. Vieira. 2006. Junk DNA as an evolutionary force. Nature 443: 521-524.
Bostock, C. 1971. Repetitious DNA. Advances in Cell Biology 2: 153-223.
Bowler, P.J. 1975. The changing meaning of “evolution”. Journal of the History of Ideas 36: 95-114.
Bremmerman, H.J. 1987. The adaptive significance of sexuality. In The Evolution of Sex and its Consequences (ed. S.C. Stearns), pp. 135-161. Birkhauser Verlag, Basel.
Brenner, S. 1998. Refuge of spandrels. Current Biology 8: R669.
Britten, R.J. and E.H. Davidson. 1969. Gene regulation for higher cells: a theory. Science 165: 349-357.
Britten, R.J. and E.H. Davidson. 1971. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Quarterly Review of Biology 46: 111-138.
Castillo-Davis, C.I. 2005. The evolution of noncoding DNA: how much junk, how much func? Trends in Genetics 21: 533-536.
Comeron, J.M. 2001. What controls the length of noncoding DNA? Current Opinion in Genetics & Development 11: 652-659.
Comings, D.E. 1972. The structure and function of chromatin. Advances in Human Genetics 3: 237-431.
Dawkins, R. 1999. The “information challenge”: how evolution increases information in the genome. Skeptic 7: 64-69.
Doolittle, W.F. and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601-603.
Doolittle, W.F. 1982. Selfish DNA after fourteen months. In Genome Evolution (eds. G.A. Dover and R.B. Flavell), pp. 3-28. Academic Press, New York.
Gall, J.G. 1981. Chromosome structure and the C-value paradox. Journal of Cell Biology 91: 3s-14s.
Georgiev, G.P. 1969. On the structural organization of operon and the regulation of RNA synthesis in animal cells. Journal of Theoretical Biology 25: 473-490.
Gibbs, W.W. 2003. The unseen genome: gems among the junk. Scientific American 289(5): 46-53.
Gibson, L.J. 1994. Pseudogenes and origins. Origins 21: 91-108.
Gilbert, W. 1978. Why genes in pieces? Nature 271: 501.
Gould, S.J. 1996. Full House. Harmony Books, New York.
Gould, S.J. 2002. The Structure of Evolutionary Theory. Harvard University Press, Cambridge, MA.
Halligan, D.L. and P.D. Keightley. 2006. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Research 16: 875-884.
Hinegardner, R. 1976. Evolution of genome size. In Molecular Evolution (ed. F.J. Ayala), pp. 179-199. Sinauer Associates, Inc., Sunderland.
Hutchinson, J., R.K.J. Narayan, and H. Rees. 1980. Constraints upon the composition of supplementary DNA. Chromosoma 78: 137-145.
Jacq, C., J.R. Miller, and G.G. Brownlee. 1977. A pseudogene structure in 5S DNA of Xenopus laevis. Cell 12: 109-120.
Jain, H.K. 1980. Incidental DNA. Nature 288: 647-648.
Jerlstrà¶m, P. 2000. Pseudogenes: are they non-functional? Creation Ex Nihilo Technical Journal 14: 15.
Kidwell, M.G. and D.R. Lisch. 2001. Transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1-24.
Kondrashov, F.A. and E.V. Koonin. 2003. Evolution of alternative splicing: deletions, insertions and origin of functional parts of proteins from intron sequences. Trends in Genetics 19: 115-119.
Kondrashov, A.S. 2005. Fruitfly genome is not junk. Nature 437: 1106.
Lefevre, G. 1971. Salivary chromosome bands and the frequency of crossing over in Drosophila melanogaster. Genetics 67: 497-513.
Loomis, W.F. 1973. Vestigial DNA? Developmental Biology 30: F3-F4.
Makalowski, W. 2003. Not junk after all. Science 300: 1246-1247.
Moore, M.J. 1996. When the junk isn’t junk. Nature 379: 402-403.
Nowak, R. 1994. Mining treasures from ‘junk DNA’. Science 263: 608-610.
Ohno, S. 1970a. Evolution by Gene Duplication. Springer-Verlag, New York.
Ohno, S. 1970b. The enormous diversity in genome sizes of fish as a reflection of nature’s extensive experiments with gene duplication. Transactions of the American Fisheries Society 1970: 120-130.
Ohno, S. 1973. Evolutional reason for having so much junk DNA. In Modern Aspects of Cytogenetics: Constitutive Heterochromatin in Man (ed. R.A. Pfeiffer), pp. 169-173. F.K. Schattauer Verlag, Stuttgart, Germany.
Ohno, S. 1974. Chordata 1: protochordata, cyclostomata, and pisces. In Animal Cytogenetics, Vol. 4 (ed. B. John), pp. 1-92. Gebrà¼der Borntraeger, Berlin.
Ohno, S. 1982. The common ancestry of genes and spacers in the euchromatic region: omnis ordinis hereditarium a ordinis priscum minutum. Cytogenetics and Cell Genetics 34: 102-111.
Ohno, S. 1985. Dispensable genes. Trends in Genetics 1: 160-164.
Patrushev, L.I. and I.G. Minkevich. 2006. Eukaryotic noncoding DNA sequences provide genes with an additional protection against chemical mutagens. Russian Journal of Bioorganic Chemistry 32: 1068-1620.
Petsko, G.A. 2003. Funky, not junky. Genome Biology 4: 104.
Raup, D.M. 1991. Exctinction. W.W. Norton & Co., New York.
Roels, H. 1966. “Metabolic” DNA: a cytochemical study. International Review of Cytology 19: 1-34.
Ruse, M. 1996. Monad to Man. Harvard University Press, Cambridge, MA.
Shapiro, J.A. and R. von Sternberg. 2005. Why repetitive DNA is essential to genome function. Biological Reviews 80: 227-250.
Sharma, A.K. 1985. Chromosome architecture and additional elements. In Advances in Chromosome and Cell Genetics (eds. A.K. Sharma and A. Sharma), pp. 285-293. Oxford and IBH Publishing Co., New Delhi.
Slack, F.J. 2006. Regulatory RNAs and the demise of ‘junk’ DNA. Genome Biology 7: 328.
Vinogradov, A.E. 1998. Buffering: a possible passive-homeostasis role for redundant DNA. Journal of Theoretical Biology 193: 197-199.
Walker, P.M.B., W.G. Flamm, and A. McLaren. 1969. Highly repetitive DNA in rodents. In Handbook of Molecular Cytology (ed. A. Lima-de-Faria), pp. 52-66. North-Holland Publishing Co., Amsterdam.
Walkup, L.K. 2000. Junk DNA: evolutionary discards or God’s tools? Creation Ex Nihilo Technical Journal 14: 18-30.
Wickelgren, I. 2003. Spinning junk into gold. Science 300: 1646-1649.
Wieland, C. 1994. Junk moves up in the world. Creation Ex Nihilo Technical Journal 8: 125.
Woodmorappe, J. 2000. Are pseudogenes ‘shared mistakes’ between primate genomes? Creation Ex Nihilo Technical Journal 14: 55-71.
Woolfe, A., M. Goodson, D.K. Goode, P. Snell, G.K. McEwen, T. Vavouri, S.F. Smith, P. North, H. Callaway, K. Kelly, K. Walter, I. Abnizova, W. Gilks, Y.J.K. Edwards, J.E. Cooke, and G. Elgar. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biology 3: e7.
Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, Libri D, Jacquier A. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725-37
Yunis, J.J. and W.G. Yasmineh. 1971. Heterochromatin, satellite DNA, and cell function. Science174: 1200-1209.
Zuckerkandl, E. 1976. Gene control in eukaryotes and the C-value paradox: “Excess” DNA as an impediment to transcription of coding sequences. Journal of Molecular Evolution 9: 73-104.
Zuckerkandl, E. and W. Hennig. 1995. Tracking heterochromatin. Chromosoma 104: 75-83.
Zuckerkandl, E. 1997. Junk DNA and sectorial gene expression. Gene 205: 323-343.
References from pop-science resources
Gregory, T. R. 2007. Genomicron Junk DNA summary
Brenner, S. 1998. Refuge of spandrels. Current Biology 8: R669.
Possibly slightly misleading article at ScientificAmerican on Junk DNA <— Read this one only after you’ve read the other ones plus my summary, otherwise you might be a bit confused!
Sandwalk has many more articles on the topic. They’re not a must-read, though, merely more information on the same.
Findandpea have another great review of the way the media failed to report properly on this. But again, scientists fell for it too, so reporters are not exclusively to blame.
Finally, Genomicron maintains an updated list of posts on the topic over at his blog. In case you’re missing anything, it can either be found at Sandwalk or at Genomicron.